When Computing You Are Allowed to Cheat

Throughout the Hydra Chronicles we have explored why group statistics are problematic in R and how to optimize their calculation. R calls are slow, but normally this is not a problem because R will quickly hand off to native routines to do the heavily lifting. With group statistics there must be at least one R call per group. When there are many groups as in the 10MM row and ~1MM group data set we’ve been using, the R-level overhead adds up.
Both data.table and dplyr resolve the problem by providing native routines
that can compute per-group statistics completely in native code for a limited
number of statistics. This relies on the group expression being in the form
statistic(variable) so that the packages may recognize the attempt to compute
known group statistics and substitute the native code. There is a more complete
discussion of this “Gforce” optimization in an earlier post. We can see
it an action here:
library(data.table)
setDTthreads(1) # for more stable timings
sys.time({ # wrapper around sytem.time; see appendix
DT <- data.table(x, grp)
var.0 <- DT[, .(var=var(x)), keyby=grp]
}) user system elapsed
1.890 0.024 1.923sys.time is a wrapper around system.time defined in the
appendix.
var.0 grp var
1: 1 0.0793
2: 2 0.0478
---
999952: 999999 0.1711
999953: 1000000 0.0668But what if we wanted the plain second central moment instead of the
unbiased estimate produced by var:
E[(X−E[X])2]E[(X−E[X])2]
Let’s start with the obvious method1:
sys.time({
DT <- data.table(x, grp)
var.1 <- DT[, .(var=mean((x - mean(x)) ^ 2)), keyby=grp]
}) user system elapsed
4.663 0.049 4.728 var.1 grp var
1: 1 0.0721
2: 2 0.0409
---
999952: 999999 0.1466
999953: 1000000 0.0594“Gforce” does not work with more complex expressions, so this ends up substantially slower. We can with some care break up the calculation into simple steps so that “Gforce” can be used, but this requires a group-join-group dance. There is an alternative: as Michael Chirico points out, sometimes we can reformulate a statistic into a different easier to compute form.
With the algebraic expansion:
(x−y)2=x2−2xy+y2(x−y)2=x2−2xy+y2
And cc a constant (scalar), XX a variable (vector), E[X]E[X] the expected value (mean) of XX, and the following identities:
E[c]=cE[E[X]]=E[X]E[cX]=cE[X]E[X+Y]=E[X]+E[Y]E[c]=cE[E[X]]=E[X]E[cX]=cE[X]E[X+Y]=E[X]+E[Y]
We can reformulate the variance calculation:
Var(X)=E[(X−E[X])2]=E[X2−2XE[X]+E[X]2]=E[X2]−2E[X]E[X]+E[X]2=E[X2]−2E[X]2+E[X]2=E[X2]−E[X]2Var(X)=E[(X−E[X])2]=E[X2−2XE[X]+E[X]2]=E[X2]−2E[X]E[X]+E[X]2=E[X2]−2E[X]2+E[X]2=E[X2]−E[X]2
The critical aspect of this reformulation is that there are no interactions between vectors and grouped values (statistics) so we can compute all the component statistics in a single pass. Let’s try it:
sys.time({
DT <- data.table(x, grp)
DT[, x2:=x^2]
var.2 <- DT[,
.(ux2=mean(x2), ux=mean(x)), keyby=grp # grouping step
][,
.(grp, var=ux2 - ux^2) # final calculation
]
}) user system elapsed
1.159 0.114 1.277 all.equal(var.1, var.2)[1] TRUEWow, that’s even faster than the “Gforce” optimized “var”. There is only one
grouping step, and in that step all expressions are of the form
statistic(variable) so they can be “Gforce” optimized.
These concepts apply equally to dplyr.
Beyond Variance
While it’s utterly fascinating (to some) that we can reformulate the variance
computation, it’s of little value since both data.table and dplyr have
compiled code that obviates the need for it. Fortunately this method is
generally applicable to any expression that can be expanded and decomposed as we
did with the variance. For example with a little elbow grease we can
reformulate the third moment:
Skew(X)=E[(X−E[X])3]=E[X3−3X2E[X]+3XE[X]2−E[X]3]=E[X3]−3E[X2]E[X]+3E[X]E[X]2−E[X]3=E[X3]−3E[X2]−2E[X]3Skew(X)=E[(X−E[X])3]=E[X3−3X2E[X]+3XE[X]2−E[X]3]=E[X3]−3E[X2]E[X]+3E[X]E[X]2−E[X]3=E[X3]−3E[X2]−2E[X]3
And to confirm:
X <- runif(100)
all.equal(
mean((X - mean(X))^3),
mean(X^3) - 3 * mean(X^2) * mean(X) + 2 * mean(X) ^3
)[1] TRUESimilarly, and what set me off on this blogpost, it is possible to reformulate the slope of a single variable linear regression slope. What Michael Chirico saw that was not immediately obvious to me is that:
Slope(X,Y)=Cov(X,Y)Var(X)=E[(X−E[X])(Y−E[Y])]E[(X−E[X])2]=E[XY]−E[X]E[Y]E[X2]−E[X]2Slope(X,Y)=Cov(X,Y)Var(X)=E[(X−E[X])(Y−E[Y])]E[(X−E[X])2]=E[XY]−E[X]E[Y]E[X2]−E[X]2
In data.table-speak:
DT <- data.table(x, y, xy=x*y, x2=x^2, grp)
slope.dt.re <- DT[,
.(ux=mean(x), uy=mean(y), uxy=mean(xy), ux2=mean(x2)),
keyby=grp
][,
.(grp, slope=(uxy - ux*uy)/(ux2 - ux^2))
] user system elapsed
1.377 0.126 1.507As with all the reformulations we’ve seen the key feature of this one is
there is no interaction between grouped statistics and ungrouped ones, allowing
a single grouping step. With the reformulation data.table is able to
beat the cumsum based base-R method we studied in an earlier
post:
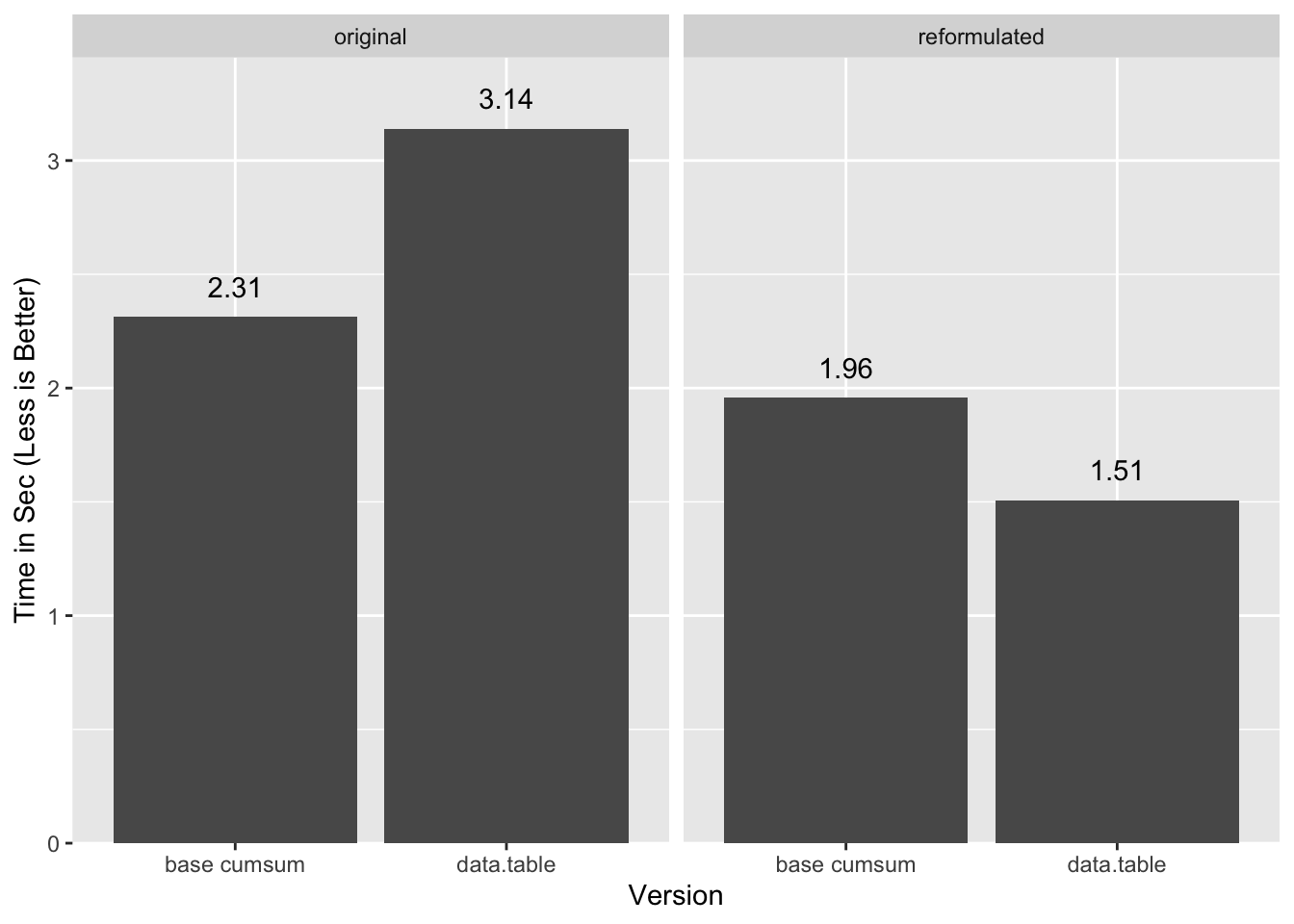
“Original” in this case is the single-pass cumsum method for base
and the group-join-group method for data.table.
More generally we should be able to apply this expansion to any nested
statistics involving sum or mean and possibly other functions, which will
cover a broad range of interesting statistics. In the case of sum, keep in
mind that:
n∑ic=ncn∑ic=nc
And similarly:
n∑i(c+X)=nc+n∑iXn∑i(c+X)=nc+n∑iX
Slow Down Cowboy
Before we run off and reformulate all the things, it is worth noting that the wikipedia page I took the reformulation of the variance from also provides a scary warning:
This equation should not be used for computations using floating point arithmetic because it suffers from catastrophic cancellation if the two components of the equation are similar in magnitude.
Uh-oh. Let’s see:
quantile(((var.2 - var.3) / var.2), na.rm=TRUE) 0% 25% 50% 75% 100%
-2.09e-07 -5.91e-16 0.00e+00 5.94e-16 2.91e-09We did lose some precision, but certainly nothing catastrophic. So under what circumstances do we actually need to worry? Let’s cook up an example that actually causes a problem, as with these values at the edge of precision of double precision numbers.
X <- c(2^52 + 1, 2^52 - 1)
mean(X)[1] 4.5e+15mean((X - mean(X))^2) # "normal" calc[1] 1mean(X^2) - mean(X)^2 # "reformulated" calc[1] 0If we carry out the algebraic expansion of the first element of X2X2 we can see that problems crop up well before the subtraction:
(X1)2=(252+20)2=(252)2+2×252×20+(20)2=2104+253+20(X1)2=(252+20)2=(252)2+2×252×20+(20)2=2104+253+20
Double precision floating point numbers only have 53 bits of precision in the fraction, which means the difference between the highest power of two and the lowest one contained in a number cannot be greater than 52 without loss of precision. Yet, here we are trying to add 2020 to a number that contains 21042104! This is completely out of the realm of what double precision numbers can handle:
identical(
2^104 + 2^53,
2^104 + 2^53 + 2^0
)[1] TRUEOne might think, “oh, that’s just the last term, it doesn’t matter”, but it does:
E[X2]=1N∑X2=12((252+20)2+(252−20)2)=12(((2104+253+20))+((2104−253+20)))=2104+20
The contributions from the middle terms cancel out, leaving only the last term as the difference between E[X2] and E[X]2.
In essence squaring of X values is causing meaningful precision to overflow out of the double precision values. By the time we get to the subtraction in E[X2]−E[X]2 the precision has already been lost. While it is true that E[X2] and E[X]2 are close in the cases of precision loss, it is not the subtraction that is the problem.
Generally we need to worry when |E[X]|≫Var(X). The point at which catastrophic precision loss happens will be around |E[X]|/Var(X)≈26, as that is when squaring X values will cause most or all the variance precision to overflow. We can illustrate with a sample with n=100, Var(X)=1, and varying ratios of |E[X]|/Var(X), repeating the experiment 100 times for each ratio:
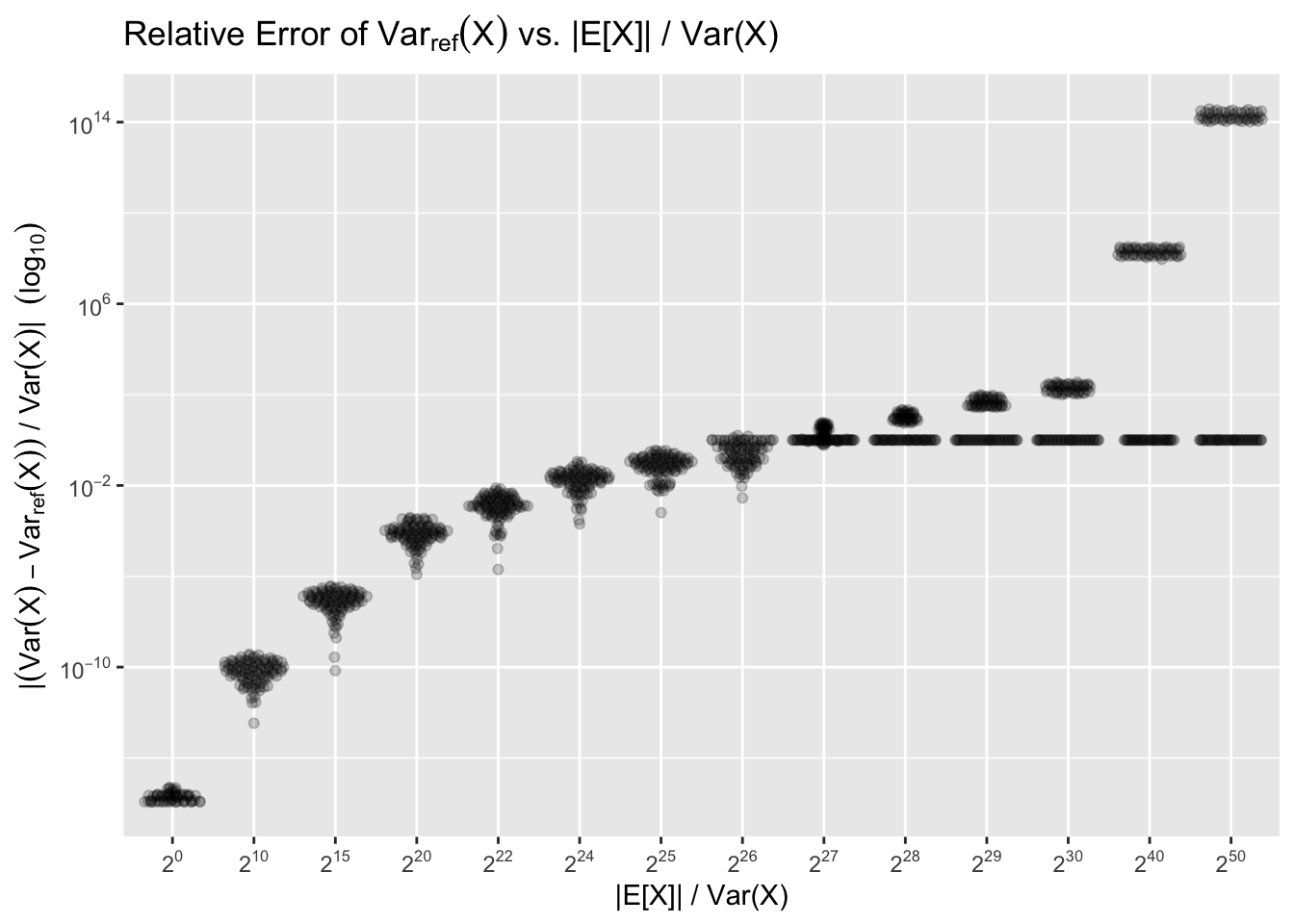
The flat lines that start forming around 226 correspond to when overflow causes the reformulation to “round down” to zero. The more dramatic errors correspond to the last bit(s) rounding up instead of down2. As the last bits are further and further in magnitude from the true value, the errors get dramatic.
If we know what the true |E[X]|/Var(X) ratio is, we can estimate the likely loss of precision. Unfortunately we cannot get a correct measure of this value from the reformulated calculations. For example, if the true ratio is 250, due to the possibility for gross overestimation the measured ratio might be as low as 22:
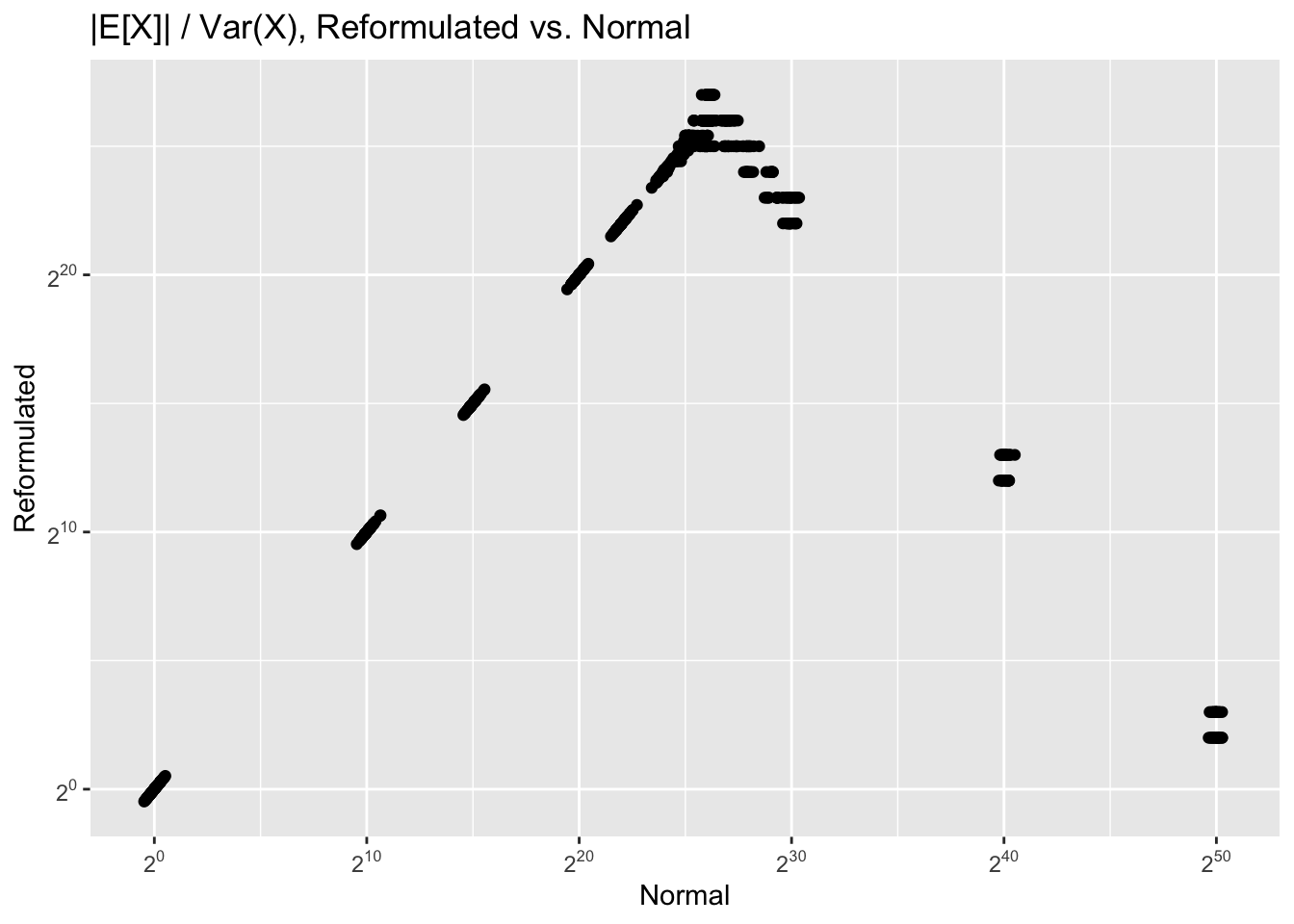
Infinite values (i.e. zero measured variance) are removed from the plot.
Another way to look at the precision loss is to examine the underlying binary representation of the floating point numbers. We do so here for each of the 100 samples of each of the attempted |E[X]|/Var(X) ratios:
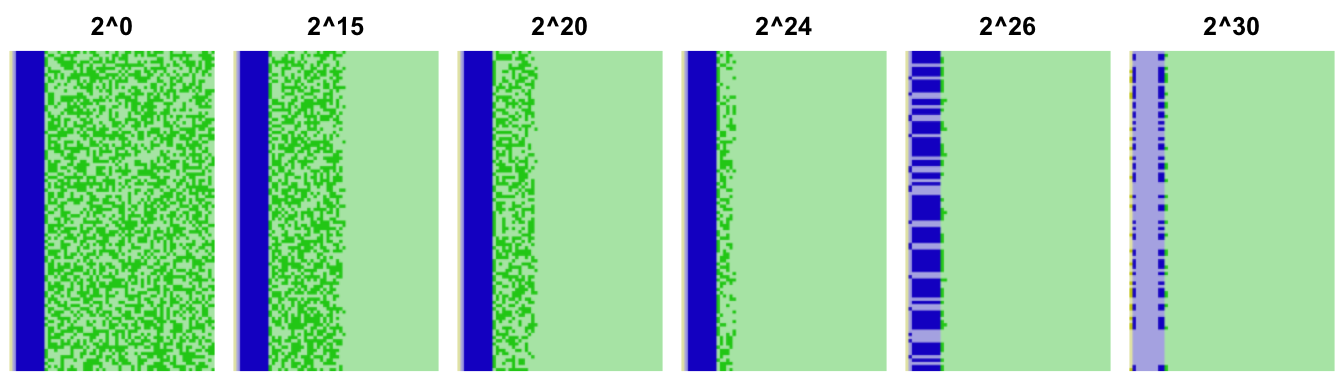
The fraction of the numbers is shown in green, and the exponent in blue. Dark
squares are ones and dark ones zeroes. The precision loss shows up as trailing
zeroes in those numbers (light green). This view makes it obvious that there is
essentially no precision by the time we get to |E[X]|/Var(X)≈26, which is why the errors in the reformulation start getting so
wild. We can estimate a lower bound on the precision by inspecting the bit
pattern, but since there may be legitimate trailing zeroes this will likely be
an under-estimate. See the appendix for a method that
checks a number has at least n bits of precision.
Each reformulation will bring its own precision challenges. The skew calculation will exhibit the same overflow problems as the variance, except further exacerbated by the cube term. The slope reformulation introduces another wrinkle due to the division of the covariance by the variance. We can illustrate with variables X and Y each with true variance 1, and means 226 and 24 respectively. We sampled from these 100 values, 100 times, and plot the resulting bit patterns for the resulting finites slopes:
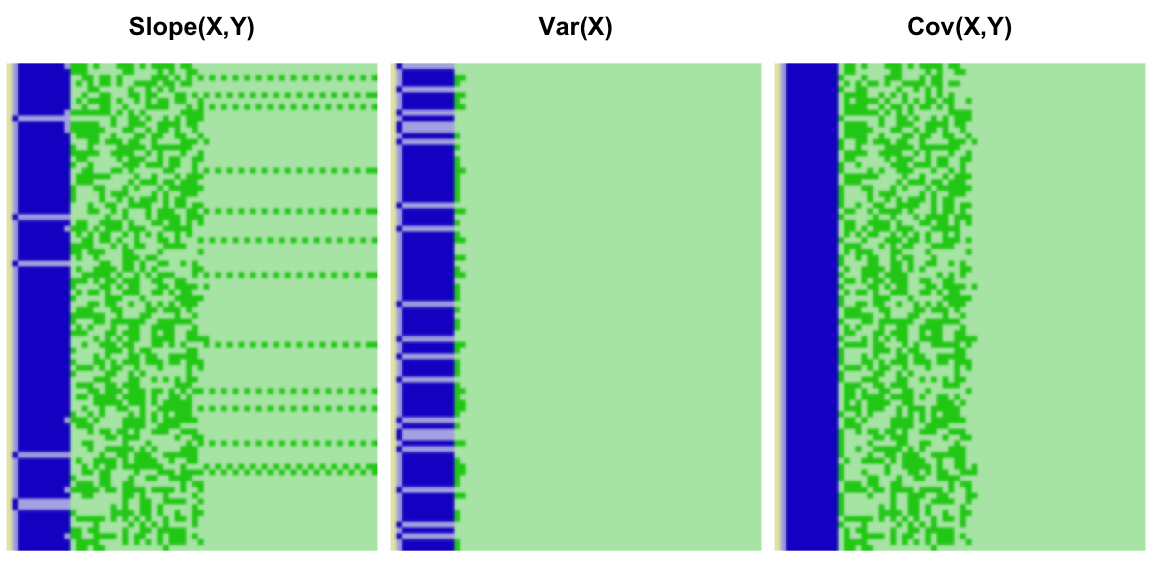
The precision of the overall calculation should be limited by that of the variance, but the interaction between the variance and covariance in all cases causes the bit pattern to overstate the available precision. In some cases, it even gives the illusion that all 53 bits are in use.
Unlike with our cumsum based group sum, I have not found a good way to
estimate a bound on precision loss. The bit pattern method is likely our best
bet, yet it feels… shaky. Not to mention that for large enough group counts
it will produce false positives as some groups will randomly have statistics
with many legitimate trailing zeroes.
A saving grace might be that, e.g. for the variance, data for which |E[X]|≫Var(X) should be reasonably rare. In cases where that might hold true, we will often have a rough order-of-magnitude estimate of E[X] which we can subtract from the data prior to computation, though this is only useful if the mean is roughly the same across all groups3.
Conclusions
Reformulations is a powerful tool for dealing with group statistics, but the precision issues are worrying because they are difficult to measure, and may take new forms with each different reformulation.
Appendix
Acknowledgments
Special thanks to Michael Chirico for making me see what should have been obvious from the get go. The best optimization is always a better algorithm, and I was too far removed from my econ classes to remember the expected value identities. So far removed in fact that my first reaction on seeing that his calculations actually produced the same result was “how is that even possible”…
Also thank you to:
- coolbutuseless for showing me that
writeBincan write directly to a raw vector. - Hadley Wickham and the
ggplot2authors forggplot2, as well as Erik Clarke and Scott Sherrill-Mix forggbeeswarm, with which I made several plots in this post. - And to all the other developers that make this blog possible.
Session Info
sessionInfo()R version 3.6.0 (2019-04-26)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS Mojave 10.14.6
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] data.table_1.12.2 dplyr_0.8.3
loaded via a namespace (and not attached):
[1] Rcpp_1.0.1 magrittr_1.5 usethis_1.5.0 devtools_2.0.2
[5] tidyselect_0.2.5 pkgload_1.0.2 R6_2.4.0 rlang_0.4.0
[9] tools_3.6.0 pkgbuild_1.0.3 sessioninfo_1.1.1 cli_1.1.0
[13] withr_2.1.2 remotes_2.0.4 assertthat_0.2.1 rprojroot_1.3-2
[17] digest_0.6.19 tibble_2.1.3 crayon_1.3.4 processx_3.3.1
[21] purrr_0.3.2 callr_3.2.0 fs_1.3.1 ps_1.3.0
[25] testthat_2.1.1 memoise_1.1.0 glue_1.3.1 compiler_3.6.0
[29] pillar_1.4.1 desc_1.2.0 backports_1.1.4 prettyunits_1.0.2
[33] pkgconfig_2.0.2 Code
dplyr Reformulate
We can apply the reformulation to dplyr, but the slow grouping holds it
back.
sys.time({
var.2a <- tibble(x, grp) %>%
mutate(x2=x^2) %>%
group_by(grp) %>%
summarise(ux2=mean(x2), ux=mean(x)) %>%
mutate(var=ux2 - ux^2) %>%
select(grp, var)
}) user system elapsed
11.171 0.299 11.660 all.equal(as.data.frame(var.2), as.data.frame(var.2a))[1] TRUEData
RNGversion("3.5.2"); set.seed(42)
n <- 1e7
n.grp <- 1e6
grp <- sample(n.grp, n, replace=TRUE)
noise <- rep(c(.001, -.001), n/2) # more on this later
x <- runif(n) + noise
y <- runif(n) + noise # we'll use this latersys.time
# Run `system.time` `reps` times and return the timing closest to the median
# timing that is faster than the median.
sys.time <- function(exp, reps=11) {
res <- matrix(0, reps, 5)
time.call <- quote(system.time({NULL}))
time.call[[2]][[2]] <- substitute(exp)
gc()
for(i in seq_len(reps)) {
res[i,] <- eval(time.call, parent.frame())
}
structure(res, class='proc_time2')
}
print.proc_time2 <- function(x, ...) {
print(
structure(
x[order(x[,3]),][floor(nrow(x)/2),],
names=c("user.self", "sys.self", "elapsed", "user.child", "sys.child"),
class='proc_time'
) ) }Check Bit Precision
The following function will check whether doubles have at least bits precision
as implied by their underlying binary encoding. The precision is defined by the
least significant “one” present in the fraction component of the number. There
is no guarantee that the least significant “one” correctly represents the
precision of the number. There may be trailing zeroes that are significant, or
a number may gain random trailing noise that is not a measure of precision.
While this implementation is reasonably fast, it is memory inefficient. Improving it to return the position of the last “one” bit will likely make it even more memory inefficient. I have only very lightly tested it so consider it more a proof of concept than a correct or robust implementation.
# requires double `x` of length > 0, and positive scalar `bits`
# assumes little endian byte order. Not tested with NA or infinite values.
has_precision_bits <- function(x, bits) {
n <- 53 - min(c(bits-1, 53))
mask.full <- n %/% 8
mask.extra <- n %% 8
mask <- as.raw(c(
rep(255, mask.full),
if(mask.extra) 2^(mask.extra) - 1,
rep(0, 8 - (mask.full + (mask.extra > 0)))
) )
colSums(matrix(as.logical(writeBin(x, raw()) & mask), nrow=8)) > 0
}By “obvious” I mean the most obvious that will also advance the narrative in the direction I want it to go in. The most obvious one would be to multiply the result of
var(...)by.N/(.N-1)to undo the bias correction.↩I have no support for this other than it makes sense. I cannot explain why there seem to be more than one “rounding up” other than it may be an artifact of the specific values that were drawn randomly for any given level of E[X]/Var(X). Normally this is something I would chase down and pin down, but honestly I’m kinda running out of patience with this endless post series.↩
In the case where each group has means that are very different from the others this is no longer a useful correction as matching each group to its mean-estimate is costly. At that point and we might as well revert to a two pass solution.↩