If Only We Could See Code In Action

I recently got caught up trying to port a JavaScript algorithm1 into R. A direct port would have been straightforward (and slow), but I wanted to vectorize it. I had a rough idea what the algorithm did from the description and from the code, but because I did not understand the semantics exactly I wasted a lot of time on bad implementations. It wasn’t until I buckled up and figured out how to step through the JS version in my browser console and painstakingly recorded what was happening that I finally got things right.
Even though I resolved the issue the experience left me wanting for a better way of visualizing algorithms in action.
Instrumenting R Functions
One of R’s lesser known super-powers is its ability to manipulate unevaluated R code. We used this to instrument an R RPN parser, which in turn allowed us to visualize it in action. Unfortunately the instrumentation was specific to that function, and generalizing it for use with the R translation of the JS algorithm felt like too much work.
How does instrumentation work? The basic concept is to modify each top-level statement to trigger a side effect. For example if we start with:
f0 <- function() {
1 + 1
}
f0()[1] 2With some language manipulation we can get:
f1 <- function() {
{
message("I'm a side effect")
1 + 1
}
}
f1()I'm a side effect[1] 2The functions produce the same result, but the second one triggers a side
effect. In this case the side effect is to issue a message, but we could have
instead recorded variable values. In other words, the side effect is the
instrumentation. I first saw this concept in Jim Hester’s wonderful
covr package where it is used to measure test code coverage.
Of course only after my need for a generalized instrumentation tool passed, I
got the irrepressible urge to write one. So I quickly threw something together
into the watcher package. It’s experimental, limited2,
but does just enough for my purposes.
watcher ships with an implementation of the insertion sort algorithm for
demonstration purposes:
library(watcher) # you'll need to install from github for this
insert_sortfunction (x)
{
i <- 2
while (i <= length(x)) {
j <- i
while (j > 1 && x[j - 1] > x[j]) {
j <- j - 1
x[j + 0:1] <- x[j + 1:0]
}
i <- i + 1
}
x
}
<bytecode: 0x7f8d1160f070>
<environment: namespace:watcher>We can use watcher::watch to transmogrify insert_sort into its instrumented
cousin insert_sort_w:
insert_sort_w <- watch(insert_sort, c('i', 'j', 'x'))This new function works exactly like the original, except that the values of the specified variables are recorded at each evaluation step and attached to the result as an attribute:
set.seed(1220)
x <- runif(10)
x [1] 0.0979 0.2324 0.0297 0.3333 0.5672 0.4846 0.4145 0.6775 0.3080 0.5123insert_sort(x) [1] 0.0297 0.0979 0.2324 0.3080 0.3333 0.4145 0.4846 0.5123 0.5672 0.6775all.equal(insert_sort(x), insert_sort_w(x), check.attributes=FALSE)[1] TRUEWe can retrieve the recorded data from the “watch.data” attribute:
dat <- attr(insert_sort_w(x), 'watch.data')
str(dat[1:2]) # show first two stepsList of 2
$ :List of 2
..$ i: num 2
..$ x: num [1:10] 0.0979 0.2324 0.0297 0.3333 0.5672 ...
..- attr(*, "line")= int 3
$ :List of 2
..$ i: num 2
..$ x: num [1:10] 0.0979 0.2324 0.0297 0.3333 0.5672 ...
..- attr(*, "line")= int 4watcher::simplify_data combines values across steps into more accessible
structures. For example, scalars are turned into vectors with one element
per step:
dat.s <- simplify_data(dat)
head(dat.s[['.scalar']]) .id .line i j
1 1 3 2 NA
2 2 4 2 NA
3 3 5 2 2
4 4 6 2 2
5 5 10 3 2
6 6 4 3 2.id represents the evaluation step, and .line the corresponding line number
from the function.
Visualizing Algorithms
watcher provides all the data we need to visualize the algorithm, but
unfortunately that is as far as it goes. Actual visualization requires a human
to map values to aesthetics. For example, here we render the values of
x as the height of 3D glass bars. Certainly we all know that 3D bar charts
are bad, but 3D bar charts made of ray-traced dielectrics? Surely there can be
nothing better!
If you would rather mine bitcoin3 with your CPU cycles we can settle for a 2D flipbook. From a pedagogical standpoint this is probably a more effective visualization, if a bit flat in comparison:
If we’re content with just the right hand side of the boring 2D visualization it is pretty easy to create that:
# Extract x data and augment with the corresponding scalar `j` loop index
xs <- dat.s[['x']]
xs <- transform(
xs, j=dat.s[['.scalar']][['j']][.id], ix=rep_len(seq_along(x), length(val))
)
# Data for labels
labs <- reshape2::melt(dat.s[['.scalar']][c('.id', 'i', 'j')], id.var='.id')
# Plot!
library(ggplot2)
p <- ggplot(xs, aes(x=ix, y=val)) +
geom_col(aes(fill=I(ifelse(!is.na(j) & ix==j, 'red', 'grey35')))) +
geom_label(
data=labs, aes(x=value, label=variable, y=-0.25 + (variable=='j') * .125)
) +
gganimate::transition_manual(.id)This will produce an approximation of the right hand side of the flipbook (showing frame 50 here):
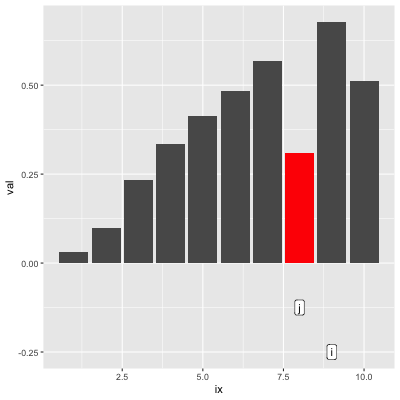
But things get tricky beyond that. Juxtaposing the code is challenging and
would benefit from some tools to render the text and graphics independently. My
own ggplot implementation requires horrid text positioning hacks.
Transitioning to the ray-traced version was relatively
easy4 thanks to the surprisingly full-featured and all-around
awesome rayrender package by Tyler Morgan Wall. I did have to
deploy my rendering farm5 though.
Conclusions
I’m unsure if watcher does enough to make visualizing algorithms more
generally practical. Perhaps automated handling of the code rendering and
juxtaposition would make this easy enough to be worth the trouble. On the
flipside, as algorithms become more complex figuring out aesthetic mappings that
intuitively represent them becomes increasingly difficult. Finally, while R
provides some great tools for instrumenting and visualizing algorithms, the
static nature of the output limits the possibilities for interactive
visualizations.
Appendix
Acknowledgments
These are post-specific acknowledgments. This website owes many additional thanks to generous people and organizations that have made it possible.
- R-core for creating and maintaining a language so wonderful it allows crazy things like self-instrumentation out of the box.
- Tyler Morgan Wall for giving us the power to bend light with
rayrender. - Jim Hester for the instrumentation concept which I borrowed from
covr; if you are interested in writing your own instrumented code please base it off of his and not mine as I just threw something together quickly with little thought. - Thomas Lin Pedersen for
gganimatewith which I prototyped some of the earlier animations, and the FFmpeg team for FFmpeg with which I stitched the frames off the ray-shaded video. - Hadley Wickham and the
ggplot2authors forggplot2with which I made many the plots in this post. - Hadley Wickham for
reshape2. - Simon Urbanek for the
pngpackage.
Adding Side Effects
So how do we turn 1 + 1 into the version with the message side effect call?
First, we create an unevaluated language template in which we can insert the
original expressions:
template <- call('{', quote(message("I'm a side effect")), NULL)
template{
message("I'm a side effect")
NULL
}We then apply this template to the body of the function:
f1 <- f0
template[[3]] <- body(f1)[[2]]
body(f1)[[2]] <- template
f1function ()
{
{
message("I'm a side effect")
1 + 1
}
}Our example function only has one line of code, but with a loop we could have just as easily modified every line of any function6.
Instrumented Insertion Sort
insert_sort_wfunction (x)
{
watcher:::watch_init(c("i", "j", "x"))
res <- {
{
.res <- (i <- 2)
watcher:::capture_data(environment(), 3L)
.res
}
while ({
.res <- (i <= length(x))
watcher:::capture_data(environment(), 4L)
.res
}) {
{
.res <- (j <- i)
watcher:::capture_data(environment(), 5L)
.res
}
while ({
.res <- (j > 1 && x[j - 1] > x[j])
watcher:::capture_data(environment(), 6L)
.res
}) {
{
.res <- (j <- j - 1)
watcher:::capture_data(environment(), 7L)
.res
}
{
.res <- (x[j + 0:1] <- x[j + 1:0])
watcher:::capture_data(environment(), 8L)
.res
}
}
{
.res <- (i <- i + 1)
watcher:::capture_data(environment(), 10L)
.res
}
}
{
.res <- (x)
watcher:::capture_data(environment(), 12L)
.res
}
}
attr(res, "watch.data") <- watcher:::watch_data()
attr(res, "watch.code") <- c("insert_sort <- function (x) ",
"{", " i <- 2", " while (i <= length(x)) {", " j <- i",
" while (j > 1 && x[j - 1] > x[j]) {", " j <- j - 1",
" x[j + 0:1] <- x[j + 1:0]", " }", " i <- i + 1",
" }", " x", "}")
res
}
<bytecode: 0x7f8d110d6390>
<environment: namespace:watcher>Session Info
sessionInfo()R version 3.6.0 (2019-04-26)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS Mojave 10.14.6
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] watcher_0.0.1 rayrender_0.4.0 ggplot2_3.2.0
loaded via a namespace (and not attached):
[1] Rcpp_1.0.1 compiler_3.6.0 pillar_1.4.2 prettyunits_1.0.2
[5] remotes_2.1.0 tools_3.6.0 testthat_2.1.1 digest_0.6.20
[9] pkgbuild_1.0.3 pkgload_1.0.2 memoise_1.1.0 tibble_2.1.3
[13] gtable_0.3.0 png_0.1-7 pkgconfig_2.0.2 rlang_0.4.0
[17] cli_1.1.0 parallel_3.6.0 withr_2.1.2 dplyr_0.8.3
[21] desc_1.2.0 fs_1.3.1 devtools_2.2.1 rprojroot_1.3-2
[25] grid_3.6.0 tidyselect_0.2.5 glue_1.3.1 R6_2.4.0
[29] processx_3.3.1 sessioninfo_1.1.1 callr_3.2.0 purrr_0.3.2
[33] magrittr_1.5 backports_1.1.4 scales_1.0.0 ps_1.3.0
[37] ellipsis_0.3.0 usethis_1.5.0 assertthat_0.2.1 colorspace_1.4-1
[41] labeling_0.3 lazyeval_0.2.2 munsell_0.5.0 crayon_1.3.4 Vladimir Agafonkin’s adaptation of the RTIN algorithm.↩
In particular recursive calls and not all control structures are supported . It should not be too difficult to add support for them, but it was not necessary for the algorithm I was working on.↩
Now that I think about it… Hey, Tyler, I have a business proposal for you 🤪…↩
Well, relatively. Obviously I had to compute positions manually and generally figure out how to use
rayrender, but I spent less time getting up to the hi-res render batch run than I did completing the ggplot2 version with the code juxtaposed.↩I had to retrieve my old 2012 11" Macbook Air to supplement my 2016 12" Macbook and manually split the frame batch across the two. Four physical cores baby! Interestingly the 2012 machine renders substantially faster. Clock speed rules sometimes; even though the 2016 machine is using a processor three generations newer, it is clocked at 1.2GHz vs 2.0 for the 2012 one. I guess it’s the price of not having a fan.↩
Control statements such as
if,for, etc. do require some additional work.↩